Methods
Assembling linked reads
First, in order to have baseline assemblies to compare against Talr, we assemble linked-reads using both LR-aware and non-LR-aware assemblers. For non-LR-aware assemblers we use ARCS to generate scaffolds using linked reads.
non-LR-aware assemblers
We assemble linked reads using three different assemblers; Unicycler, SPAdes, and ABySS. The example below shows how to do this:
make unicycler
make spades
make abyss
This will give us assembled genomes in FASTA format. In addition, we will get graph representation of the assemblies in GFA formats (or FASTG for SPAdes).
results/assemblies/f1chr4.abyss.contigs.fa
results/assemblies/f1chr4.abyss.contigs.gfa
results/assemblies/f1chr4.abyss.scaffolds.fa
results/assemblies/f1chr4.abyss.scaffolds.gfa
results/assemblies/f1chr4.spades.contigs.fa
results/assemblies/f1chr4.spades.fastg
results/assemblies/f1chr4.spades.scaffolds.fa
results/assemblies/f1chr4.unicycler.fa
results/assemblies/f1chr4.unicycler.gfa
We can visualize these graph formats using Bandage. For example, to visualize assemblies obtained by Unicycler we run:
Bandage load results/assemblies/f1chr4.unicycler.gfa
Scaffolding assemblies using ARCS
One could think of improving assemblies obtained from non-LR-aware assemblers by scaffolding them using linked reads. This can be done using ARCS which is designed specifically to scaffold draft assemblies using linked reads. These assemblies can be obtained by running the following commands:
make unicycler-arcs
make spades-arcs
make abyss-arcs
This will give us scaffolds generated by ARCS. In this example, the list of output files is as follows:
results/arcs-abyss-scaffold/f1chr4.abyss.contigs.fa
results/arcs-abyss-scaffold/f1chr4.abyss.scaffolds.fa
results/arcs-abyss-scaffold/f1chr4.spades.contigs.fa
results/arcs-abyss-scaffold/f1chr4.spades.scaffolds.fa
results/arcs-abyss-scaffold/f1chr4.unicycler.fa
LR-aware assemblers
Results
Assessment of assemblies
We can use QUAST to compare Talr with other assemblies generated above. This can be easily done using the following command:
make results/quast/quast-f1chr4/report.txt
Here is a summary of the results for chr4 of the Fruit Fly genome.
| Assembly | NG50 | Quast-misassemblies |
|---|---|---|
| ABySS | 53,005 | 4 |
| SPAdes | 77,426 | 5 |
| Unicycler | 61,681 | 5 |
| ABySS+ARCS | 936,628 | 10 |
| SPAdes+ARCS | 988,436 | 11 |
| Unicycler+ARCS | 322,017 | 9 |
| Talr+SPAdes+Miniasm | 195,672 | 6 |
| Talr+SPAdes+Miniasm (sdj) | 160,342 | 8 |
| Talr+Unicycler+Miniasm | 112,721 | 7 |
| Talr+SPAdes+Unicycler | 254,061 | 5 |
| Talr+SPAdes+Unicycler+ARCS | 1,126,501 | 6 |
Unicycler (baseline)
Assemble the linked reads using Unicycler, ignoring barcodes.
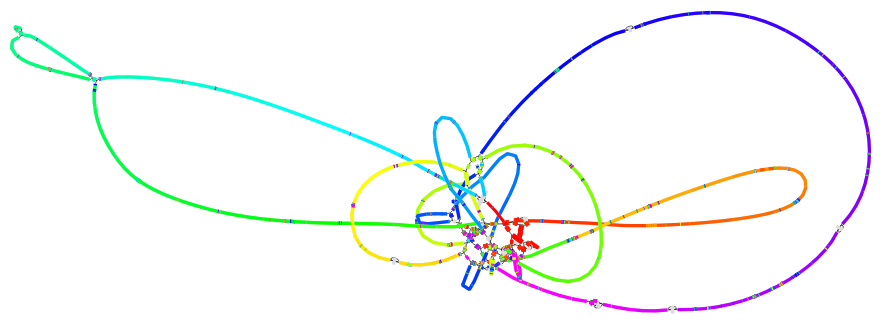
Physlr
Construct a physical map of the linked read large molecules.

Talr and SPAdes
Extract a FASTQ file containing the reads for those barcodes found in each region. Assemble the reads of each region using SPAdes.
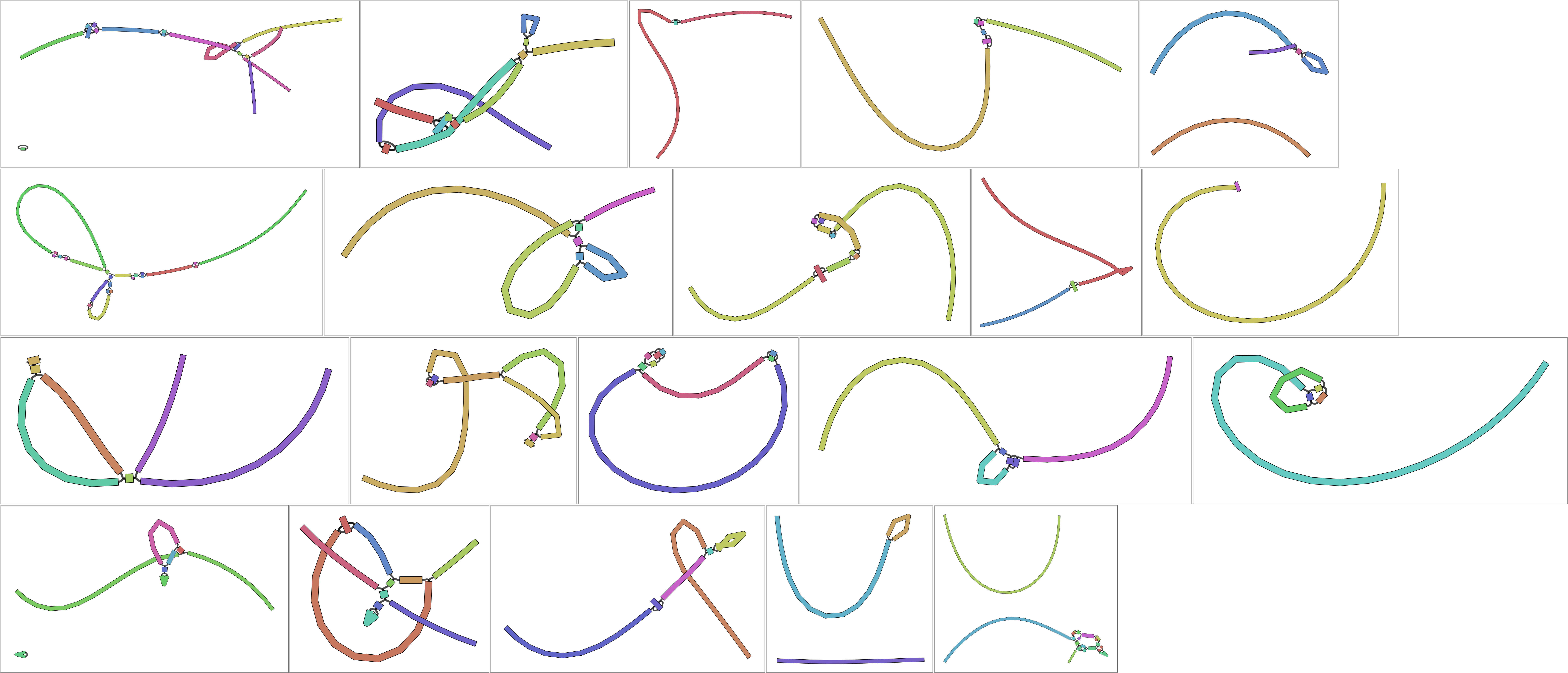
Unicycler
Assemble the short reads using Unicycler, and use all of the SPAdes contigs as “long reads”.
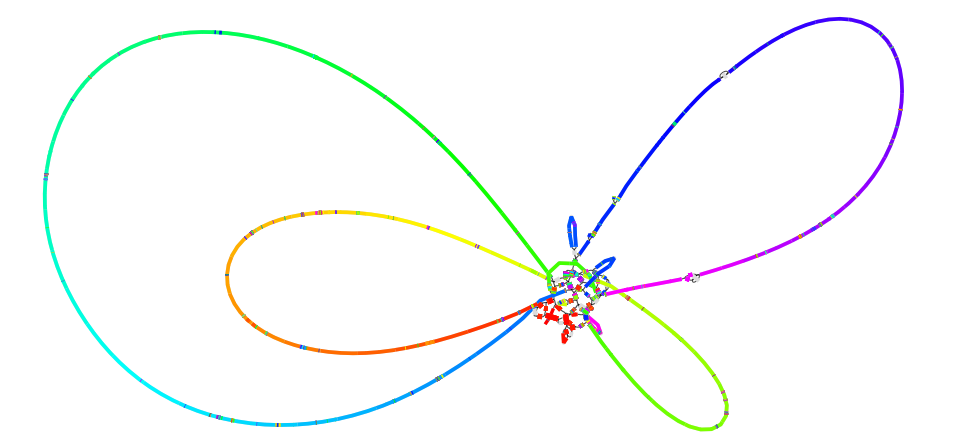
ARCS
Scaffold the Unicycler contigs with the linked reads using ARCS.