Organellar Genomes of Sitka Spruce (Picea sitchensis) Assembly and Annotation
Shaun Jackman @sjackman
Benjamin P Vandervalk, Rene L Warren, Hamid Mohamadi, Justin Chu, Sarah Yeo, Lauren Coombe, Stephen Pleasance, Robin J Coope, Joerg Bohlmann, Steven JM Jones, Inanc Birol
2017-01-15
Shaun Jackman

Background
Conifer Genomics
- Conifer genomes are large, about twenty gigabases
- Before 2013 no conifer genomes had been published
- Then three in the period of one year!
- Loblolly pine (Neale et al. 2014)
- Norway spruce (Nystedt et al. 2013)
- White spruce (Birol et al. 2013)

Genome Skimming
- Whole genome sequencing data contains both nuclear and organellar reads
- Each cell contains hundreds of mitochondria and plastids
- Reads of the organellar genomes are abundant
- Organellar sequences assemble well with a single lane
- Single-copy nuclear sequences are too low depth to assemble well
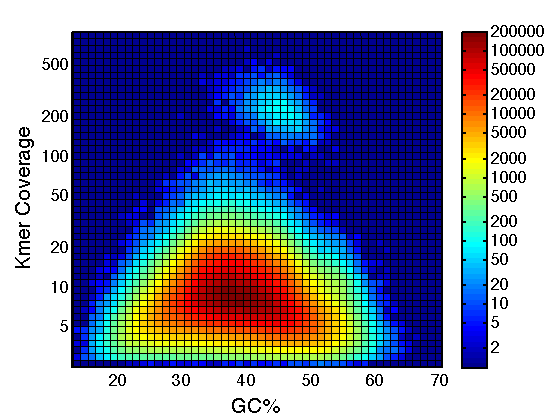
Classification
- Assembly is composed of
- plastid sequence
- mitochondrial sequence
- nuclear repeat elements
- Identify putative organellar sequences by
- homology to known organellar sequences
- depth of coverage
- length
- GC content
Work so far
- Assembled and published the annotated sequences of
complete white spruce plastid genome
draft white spruce mitochondrial genome
(Jackman et al. 2015) - Assembled and published the annotated sequence of
complete Sitka spruce plastid genome
(Coombe et al. 2016) - Currently working to assemble and annotate the
draft Sitka spruce mitochondrial genome
White Spruce Organelles
- Illumina paired-end
- One 2x300 MiSeq lane for the plastid
- One 2x150 HiSeq lane for the mitochondrion
- Illumina mate-pair for scaffolding
- Assemble with ABySS
- Complete plastid genome in one contig
- Draft mitochondrial genome in 36 scaffolds
- Close gaps with Sealer
- Polish with Pilon
- Annotate genes with Maker and Prokka
- Manual annotation of difficult genes
- Three genes with short initial exons < 10 bp
- One trans-spliced gene (rps12)

10x Genomics Chromium
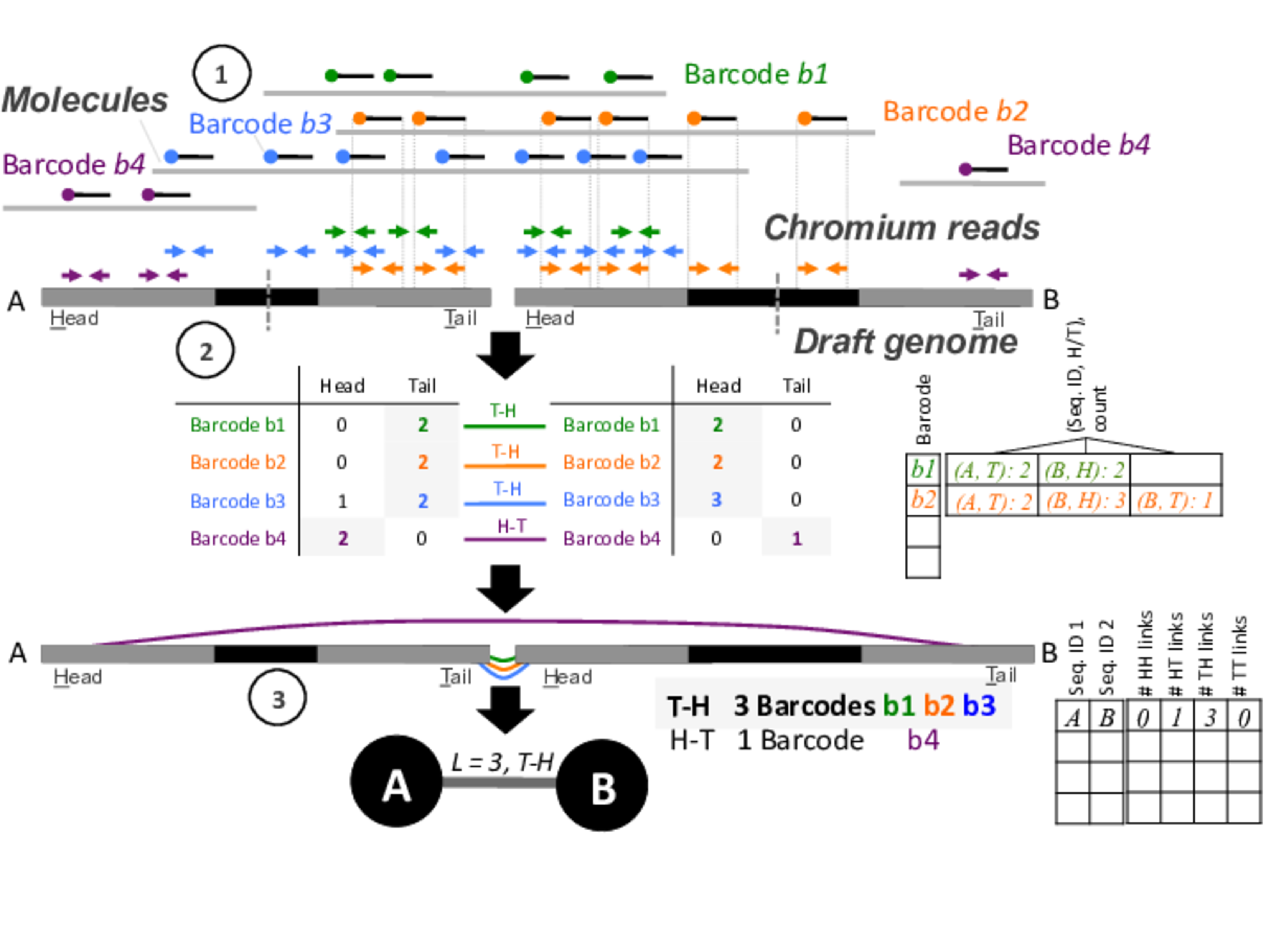
Sitka Spruce Plastid
- One lane of Illumina 2x150 HiSeq of 10x GemCode
- One library rather than two:
Illumina paired-end and mate-pair - Assemble with ABySS
- Scaffold with ARCS and LINKS
- Close gaps with Sealer
- Polish with Pilon
- Annotate with Maker
- Manual annotation of difficult genes
- Complete plastid genome in one contig
- Perfect synteny to white spruce plastid

Sitka Spruce Mitochondrion
Aim
Assemble the Sitka spruce mitochondrion into a single scaffold* using 10x Chromium data and annotate it.
* if it has a single chromosome
Method
Classification
- Assembly is composed of
- plastid sequence
- mitochondrial sequence
- nuclear repeat elements
- Identify putative organellar sequences by
- homology to known organellar sequences
- depth of coverage
- length
- GC content
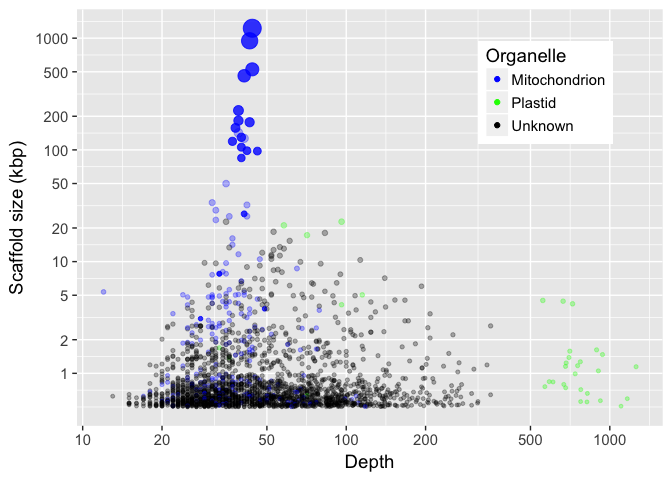
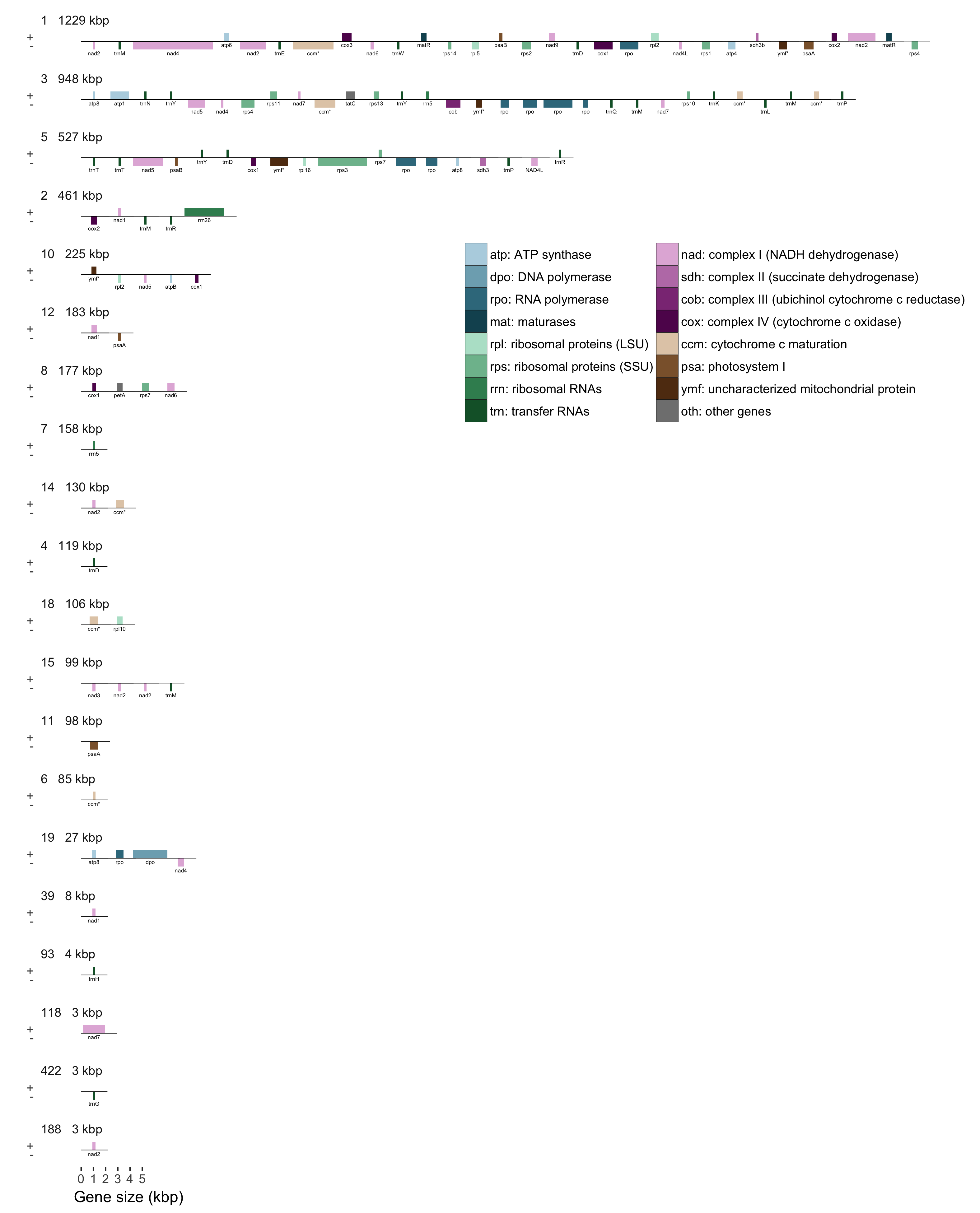
Results
- Largest scaffold is 1.2 Mbp
- 50% of the 6 Mbp genome in 4 scaffolds > 460 kbp
- 75% of the genome in 13 scaffolds > 100 kbp
- 1/223 or 0.45% of reads are mitochondrial
- 115 ORFs with similarity to known mitochondrial genes
- 1,154 other ORFS ≥ 300 bp
- 9 Type II introns in 6 genes
fin
Shaun Jackman
Slides
https://sjackman.ca/picea-sitchensis-organelles-slides
Markdown source code
https://github.com/sjackman/
picea-sitchensis-organelles-slides
Links
ABySS · ARCS · Architect · Fragscaff · LINKS · LongRanger · MAKER · Pilon · Prokka · RNAweasel · Sealer · Supernova
Supplementary Slides
Future Work
| Read Metrics | Plastid | Mitochondrion |
|---|---|---|
| Number of HiSeq lanes | 1 GemCode lane | 1 Chromium lane |
| Read length | 2 x 125 bp | 2 x 150 bp |
| Number of read | 630 million | 843 million |
| Number selected for assembly | 4.3 million | 119 million |
| Number mapped to assembly | 15,232 of 4.3 M | 3.78 M of 843 M |
| Proportion of organellar reads | 1/283 0.35% | 1/223 or 0.45% |
| Depth of coverage | 17x | 40x |
| Assembly Metrics | Plastid | Mitochondrion |
|---|---|---|
| Assembled genome size | 124,029 bp | 6.09 Mbp |
| Number of contigs | 1 contig | 1,216 contigs |
| Contig N50 | 124 kbp | 13.7 kbp |
| Number of scaffolds | 1 scaffold | 239 scaffolds |
| Scaffold N50 | 124 kbp | 461 kbp |
| Largest scaffold | 124 kbp | 1,223 kbp |
| GC content | 38.8% | 43.6% |
| Annotation Metrics | Plastid | Mitochondrion |
|---|---|---|
| Number of genes w/o ORFS | 114 (108) | 115 (67) |
| Protein-coding genes (mRNA) | 74 (72) | 84 (47) |
| rRNA genes | 4 (4) | 3 (2) |
| tRNA genes | 36 (32) | 25 (18) |
| ORFs ≥ 300 bp | 4 | 1,154 |
| Introns in coding genes | 9 (8) | 9 (6) |
| Introns in tRNA genes | 6 (6) | 0 |
