Efficient Assembly of Large Genomes
Doctoral Examination
Shaun Jackman
2019-04-16
Shaun Jackman
Efficient Assembly
of Large Genomes
- Introduction
- ABySS 2.0
- Tigmint
- UniqTag
- ORCA
- Organellar genomes of white spruce
- Mitochondrial genome of Sitka spruce
- Genome assembly of western redcedar
- Conclusion
Sitka Spruce Mitochondrion
Submitted
2019 doi.org/c4mv
![Sitka Spruce Mitochondrion]()
ORCA
Bioinformatics
2019 doi.org/c4mw
![ORCA]()
Tigmint
BMC Bioinformatics
2018 doi.org/cwfh
![Tigmint]()
Submitted
2019 doi.org/c4mv

ORCA
Bioinformatics
2019 doi.org/c4mw
Tigmint
BMC Bioinformatics
2018 doi.org/cwfh
ABySS 2.0
Genome Research
2017 doi.org/f9x8qp
![ABySS 2.0]()
White Spruce Organelles
Genome Biology and Evolution
2016 doi.org/f8bxck
![Organellar Genomes of White Spruce]()
UniqTag
PLOS ONE
2015 doi.org/c3m3
![UniqTag]()
Genome Research
2017 doi.org/f9x8qp

White Spruce Organelles
Genome Biology and Evolution
2016 doi.org/f8bxck

UniqTag
PLOS ONE
2015 doi.org/c3m3

Short Read Genome Assembly
ABySS 1.0 (2009) was the first to assemble
a human genome from short reads (42 bp!)
a human genome from short reads (42 bp!)
- de Bruijn graph assembler
- Stored k-mers in a hash table
- Distributed the hash table over many machines
- Used MPI to aggregate sufficient memory
- Assembles large genomes
Challenges
- Uses lots of memory
- Network communication is super slow
- Message passing is also slow
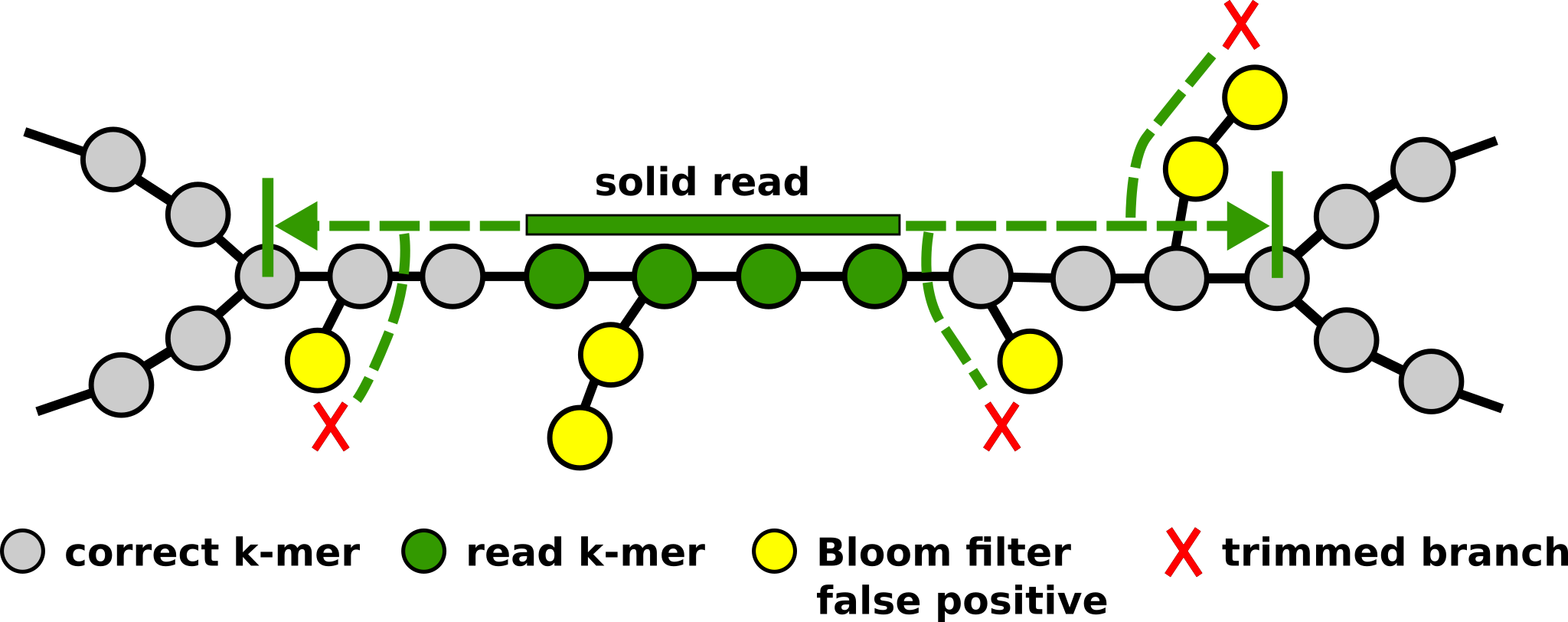
Solution
- A memory-efficient data structure
reduces memory usage - Fitting entire graph in a single machine
eliminates network communication - Using shared memory (OpenMP)
eliminates message passing (MPI)
ABySS 2.0 reduces the memory
usage of ABySS by ten fold.
usage of ABySS by ten fold.
Memory usage is independent of k

Spruce genome assemblies
| ABySS | 1.3.5 | 2.0.0 |
|---|---|---|
| Spruce species | Interior | Sitka |
| Machines | 115 | 1 |
| RAM (GB) | 4,300 | 500 |
| CPU cores | 1,380 | 64 |
| CPU time* | 6.0 years | 3.2 years |
* Time of unitig assembly without scaffolding
ABySS 2.0 Conclusions
- ABySS 2.0 reduces memory usage by 10 fold
from 418 GB to 34 GB for human
from 4,300 GB to 500 GB for spruce - High-throughput short-read sequencing
combined with large molecule scaffolding
such as linked reads and optical mapping
permits cost effective assembly of large genomes
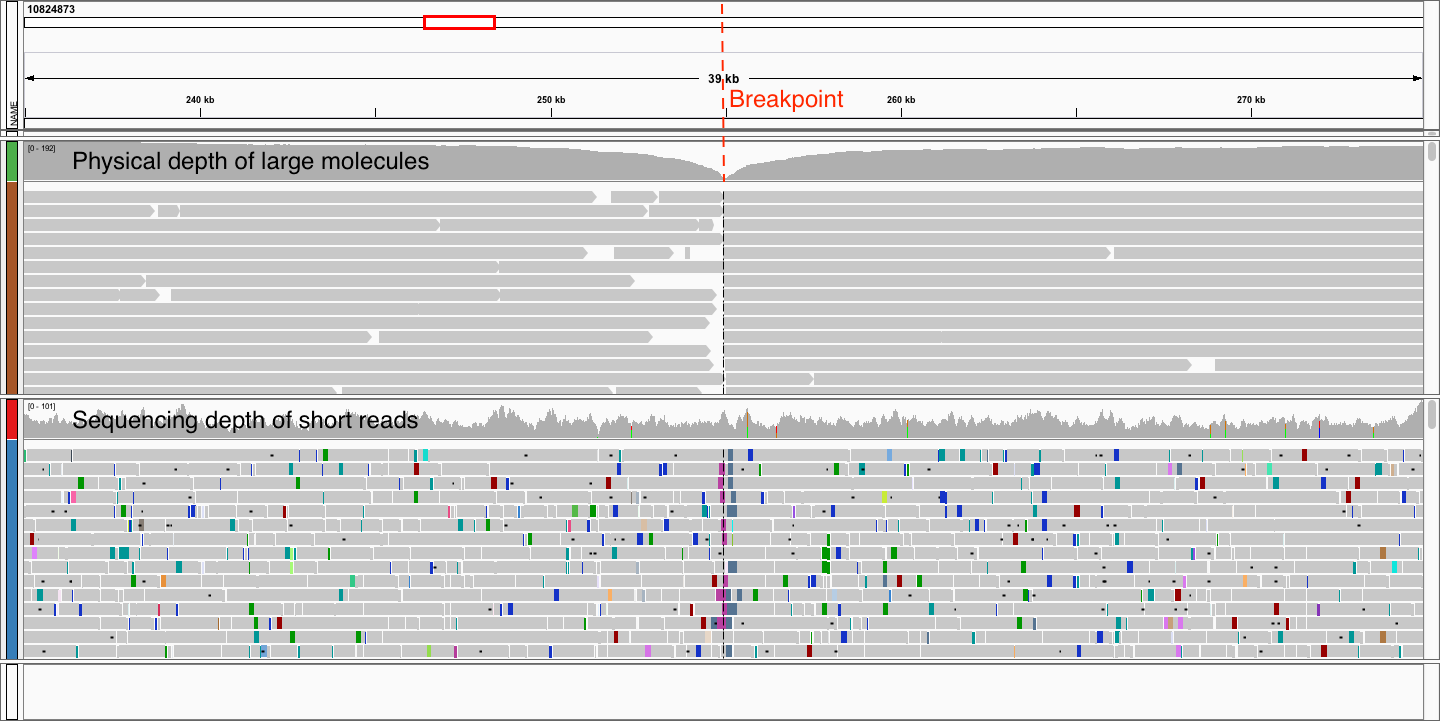
Linked Reads
Contigs and scaffolds
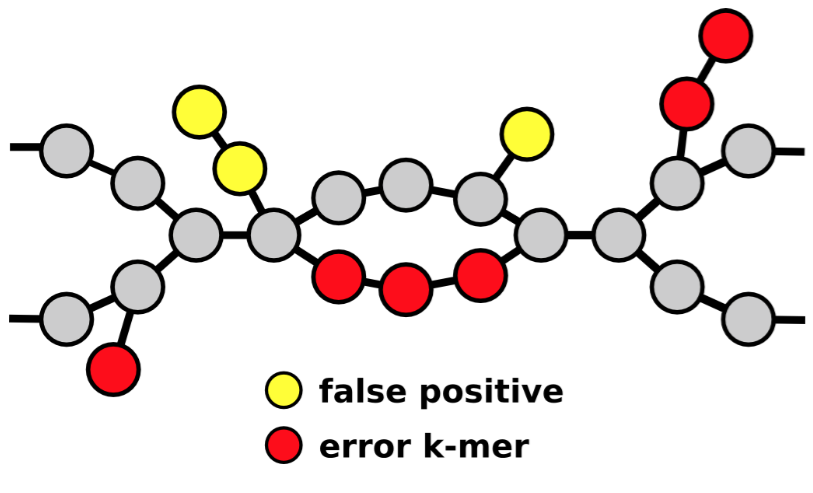
come to an end due to…
repeats
sequencing gaps
structural variation
misassemblies
sequencing gaps
structural variation
misassemblies

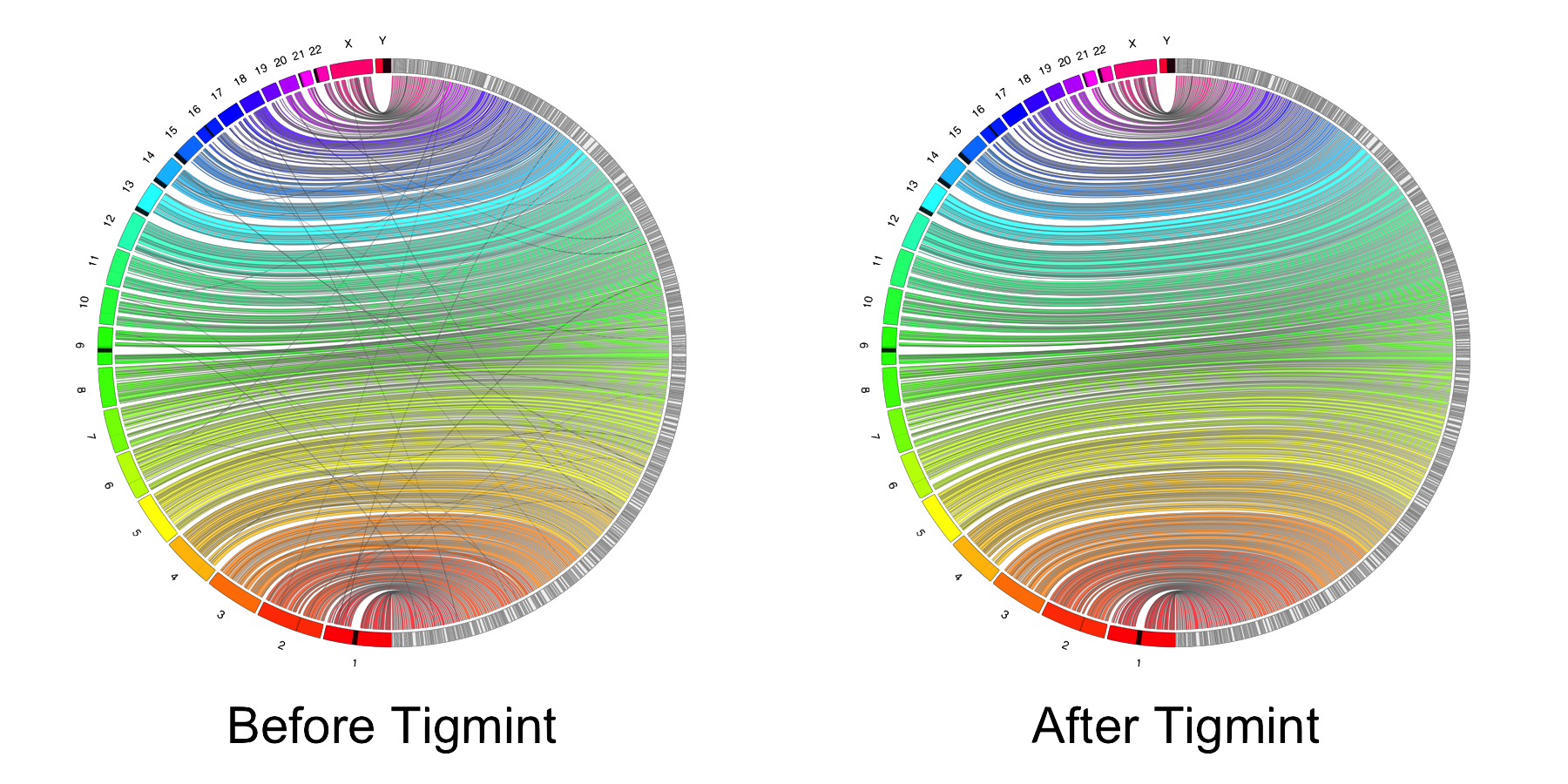
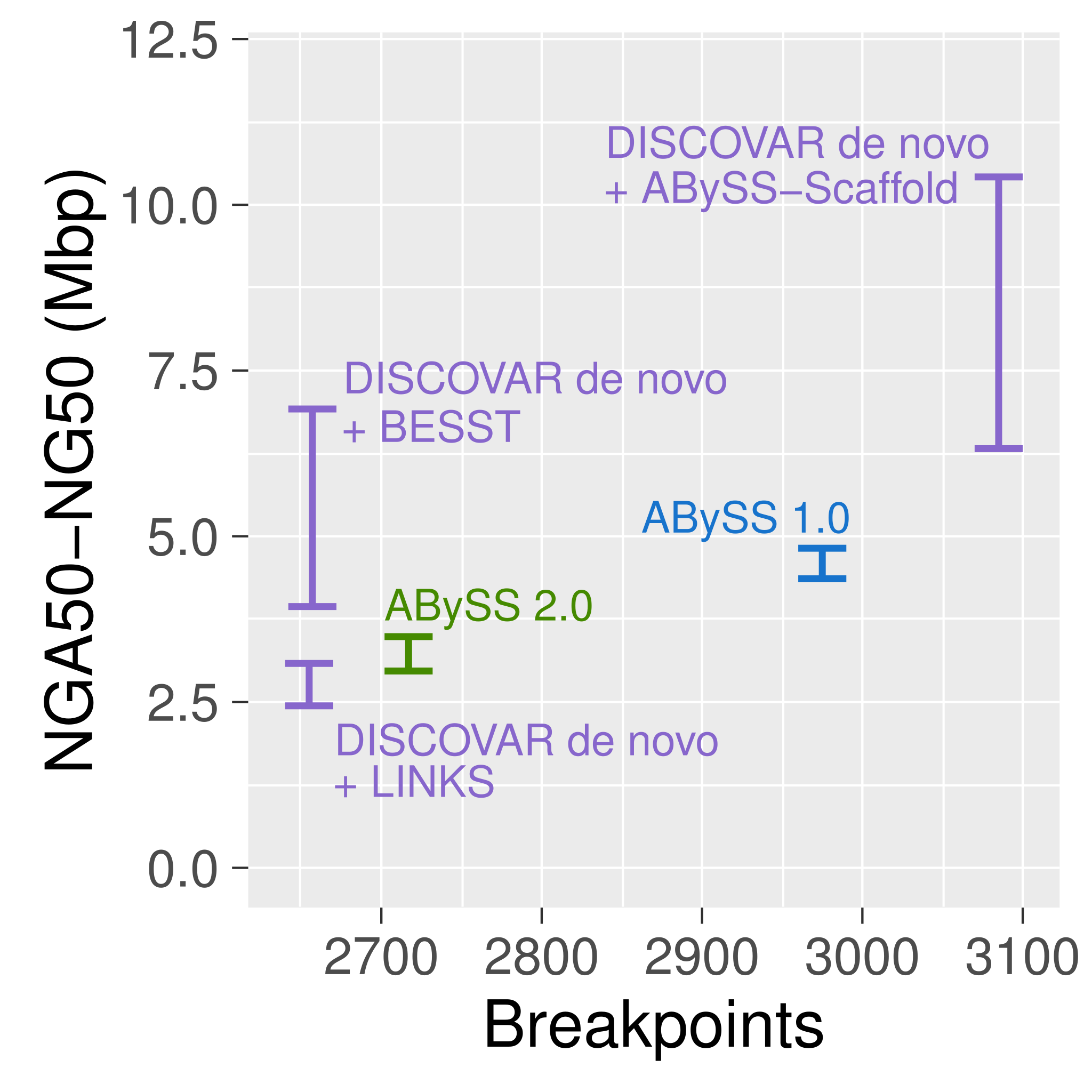
Correct misassemblies

Scaffold

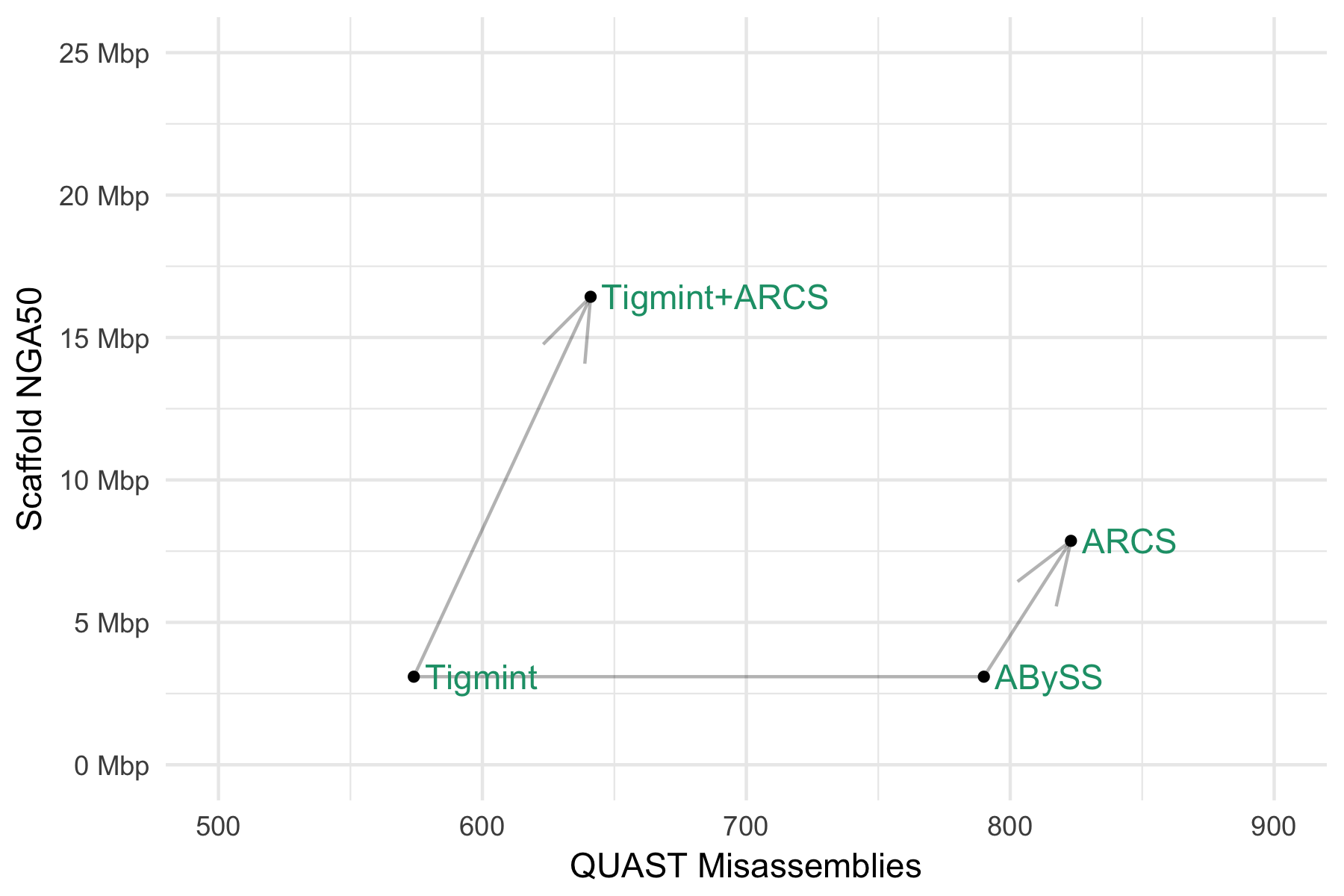
| Assembly Tools | NGA50 |
|---|---|
| ABySS 2.0 | 3 Mbp |
| ABySS 2.0 + ARCS | 8 Mbp |
| ABySS 2.0 + Tigmint + ARCS | 16 Mbp |
Tigmint reduced misassemblies by 216 (27% reduction)
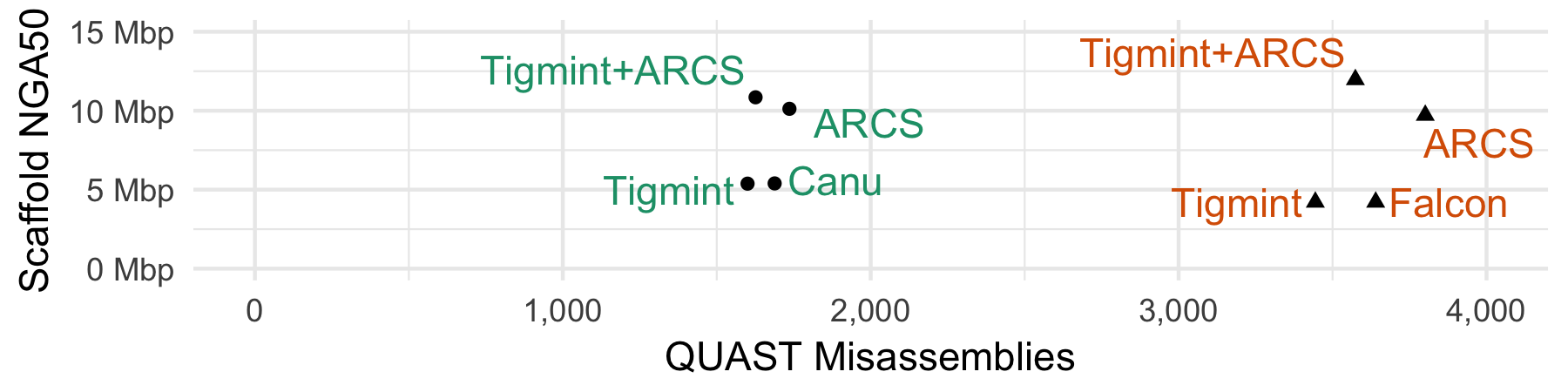
| Sequencing | Nanopore | PacBio |
|---|---|---|
| Assembler | Canu | Falcon |
| NGA50 before | 5.4 Mbp | 4.2 Mbp |
| NGA50 after | 10.9 Mbp | 12.0 Mbp |
| Improvement | 2.0 fold | 2.9 fold |
Tigmint Conclusions
Scaffolding after correcting with Tigmint yields an assembly both more correct and more contiguous
Linked reads permit cost-effective assembly of large genomes using high-throughput sequencing
Western redcedar (Thuja plicata)

Western Redcedar Methods

Conifer Assemblies
| Year | Species | Scaffold N50 |
|---|---|---|
| 2018 | Western redcedar | 2,310 kbp |
| 2017 | Sugar pine2 | 2,510 kbp |
| 2017 | Douglas fir | 341 kbp |
| 2017 | Loblolly pine2 | 108 kbp |
| 2016 | Sugar pine1 | 247 kbp |
| 2015 | Interior white spruce2 | 83 kbp |
| 2015 | White spruce | 20 kbp |
| 2014 | Loblolly pine1 | 67 kbp |
| 2013 | Interior white spruce1 | 20 kbp |
| 2013 | Norway spruce | 5 kbp |
1initial assembly 2improved assembly
Efficient Assembly
of Large Genomes
- Introduction
- ABySS 2.0 (doi.org/f9x8qp)
- Tigmint (doi.org/cwfh)
- UniqTag (doi.org/c3m3)
- ORCA (doi.org/c4mw)
in press - Organellar genomes of white spruce (doi.org/f8bxck)
- Mitochondrial genome of Sitka spruce (doi.org/c4mv)
submitted - Genome assembly of western redcedar
- Conclusion
Research Supervisors
Committee Members
University Examiners
External Examiner
C. Titus Brown, Genome Center
University of California, Davis
University of California, Davis
Chair
Jiahua Chen, Statistics
fin
Supplemental Slides
Publications
- Five first-author (or joint) papers
- One paper each year from 2015 through 2019
- One first-author manuscript submitted
(Sitka spruce mitochondrion) - Collaborated on 32 papers since 2009
- 28 papers with at least 10 citations
- ABySS has been cited over 2,700 times!
First-author Publications
- Largest Complete Mitochondrial Genome of a Gymnosperm, Sitka Spruce (Picea sitchensis), Indicates Complex Physical Structure
SD Jackman, L Coombe, RL Warren, H Kirk, E Trinh, T McLeod, S Pleasance, P Pandoh, Y Zhao, RJ Coope, J Bousquet, J Bohlmann, SJM Jones, I Birol
(submitted) 2019 - ORCA: A Comprehensive Bioinformatics Container Environment for Education and Research
SD Jackman, T Mozgacheva, S Chen, B O’Huiginn, L Bailey, I Birol, SJM Jones
Bioinformatics 2019 (in press) - Tigmint: correcting assembly errors using linked reads from large molecules
SD Jackman, L Coombe, J Chu, RL Warren, BP Vandervalk, …
BMC Bioinformatics 2018 - ABySS 2.0: resource-efficient assembly of large genomes using a Bloom filter
SD Jackman*, BP Vandervalk*, H Mohamadi, J Chu, S Yeo, SA Hammond, …
Genome Research 2017 - Organellar genomes of white spruce (Picea glauca): assembly and annotation
SD Jackman, RL Warren, EA Gibb, BP Vandervalk, H Mohamadi, J Chu, …
Genome Biology and Evolution 2015 - UniqTag: content-derived unique and stable identifiers for gene annotation
SD Jackman, J Bohlmann, I Birol
PLOS ONE 2015
Selected Publications
- Assembly of the complete Sitka spruce chloroplast… L Coombe, RL Warren, SD Jackman, C Yang, BP Vandervalk, …, I Birol
PloS one 2016 - Spaced seed data structures for de novo assembly
I Birol, J Chu, H Mohamadi, SD Jackman, K Raghavan, …, RL Warren
International journal of genomics 2015 - Konnector v2.0: pseudo-long reads from PE sequencing
BP Vandervalk, C Yang, Z Xue, K Raghavan, J Chu, H Mohamadi, SD Jackman, …, I Birol
BMC medical genomics 2015 - Sealer: a scalable gap-closing application…
D Paulino, RL Warren, BP Vandervalk, A Raymond, SD Jackman, I Birol
BMC Bioinformatics 2015 - On the representation of de Bruijn graphs
R Chikhi, A Limasset, SD Jackman, JT Simpson, P Medvedev
Journal of Computational Biology 2015 - Improved white spruce (Picea glauca) genome…
RL Warren, CI Keeling, MMS Yuen, A Raymond, GA Taylor, …, J Bohlmann
The Plant Journal 2015 - Assembling the 20Gb white spruce genome…
I Birol, A Raymond, SD Jackman, S Pleasance, R Coope, …, SJM Jones
Bioinformatics 2013
ABySS 1.0
| Human | Spruce | |
|---|---|---|
| Genome size | 3 Gbp | 20 Gbp |
| RAM | 418 GB | 4.3 TB |
| CPU cores | 64 | 1,380 |
| Wall time | 14 hours | 12 days |
| Year | 2009 & 2017 | 2013 |
| Short DOI | doi.org/f9x8qp | doi.org/f4zzrr |
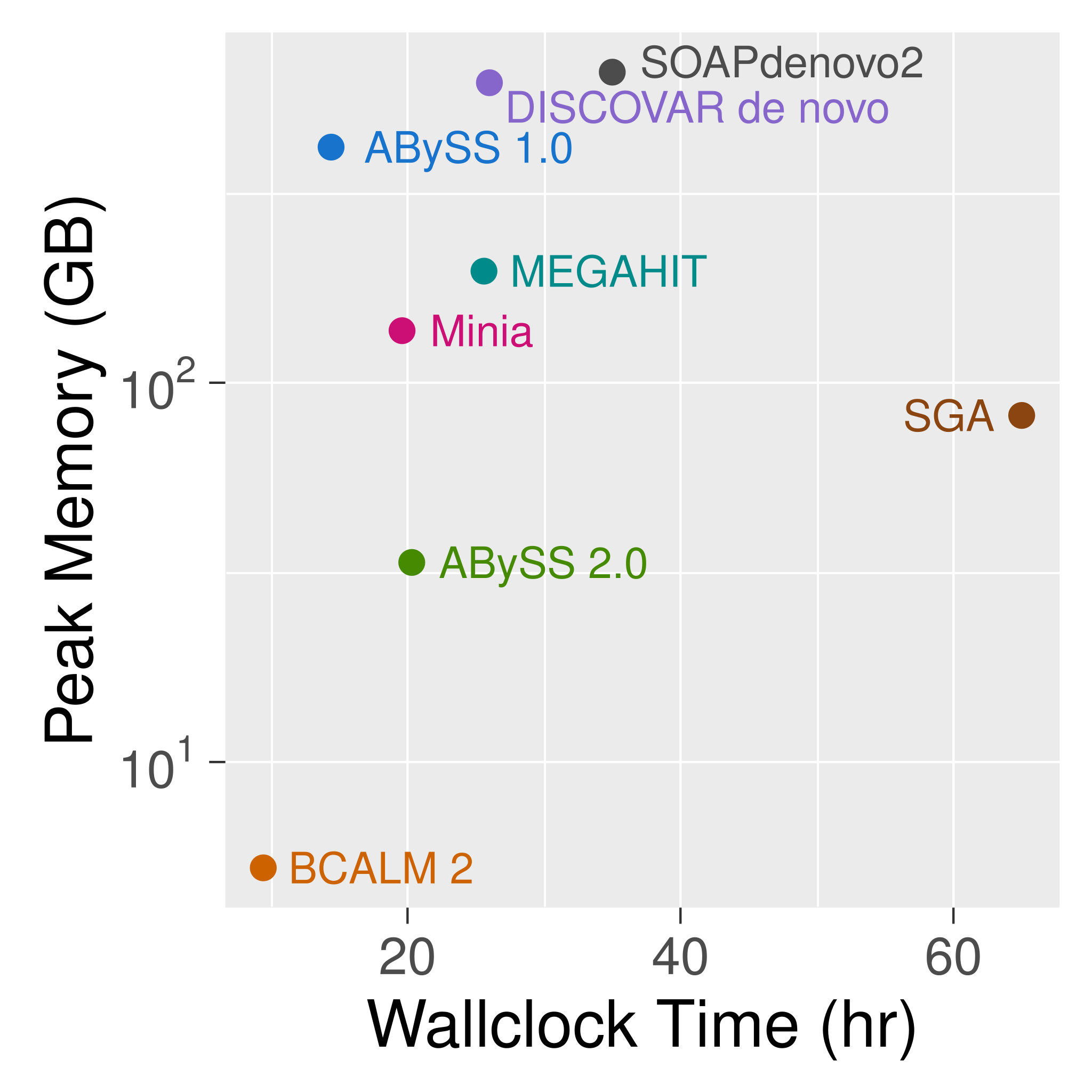
Spruce genome assemblies
| ABySS | 1.3.5 | 2.0.0 |
|---|---|---|
| Spruce species | Interior | Sitka |
| Machines | 115 | 1 |
| RAM (GB) | 4,300 | 500 |
| CPU cores | 1,380 | 64 |
| CPU time* (years) | 6.0 | 3.2 |
| Wall time* (days) | 1.6 | 18 |
| Year | 2013 | 2017 |
| Short DOI | doi:f4zzrr | NA |
* Time of unitig assembly without scaffolding
Tools for Linked Reads
Align linked reads
Lariat (Long Ranger) · EMA
Structural variants
Long Ranger · GROC-SVs · NAIBR · SVenX · Topsorter
Phase variants
Long Ranger
Genome sequence assembly
Supernova
Scaffolding
ARCS · Architect · Fragscaff · Scaff10x
Lariat (Long Ranger) · EMA
Structural variants
Long Ranger · GROC-SVs · NAIBR · SVenX · Topsorter
Phase variants
Long Ranger
Genome sequence assembly
Supernova
Scaffolding
ARCS · Architect · Fragscaff · Scaff10x
Tigmint Method
- Map reads to the assembly
- Group reads within d bp of each other (d = 50 kbp)
- Infer start and end coordinates of molecules
- Construct an interval tree of the molecules
- Each w bp region ought to be spanned by n molecules
(w = 1 kbp, n = 20) - Identify regions with fewer than n spanning molecules
- Cut sequences at regions with insufficient coverage

Note: Supernova used only linked reads, others PE+MP+LR.
Tigmint Time and Memory
bwa mem Map reads to assembly5½ hours, 17 GB RAM, 48 threads
tigmint-molecule Group reads into molecules3¼ hours, 0.08 GB RAM, 1 thread
tigmint-cut Identify misassemblies and cut sequences7 minutes, 3.3 GB RAM, 48 threads
Western Redcedar Assembly
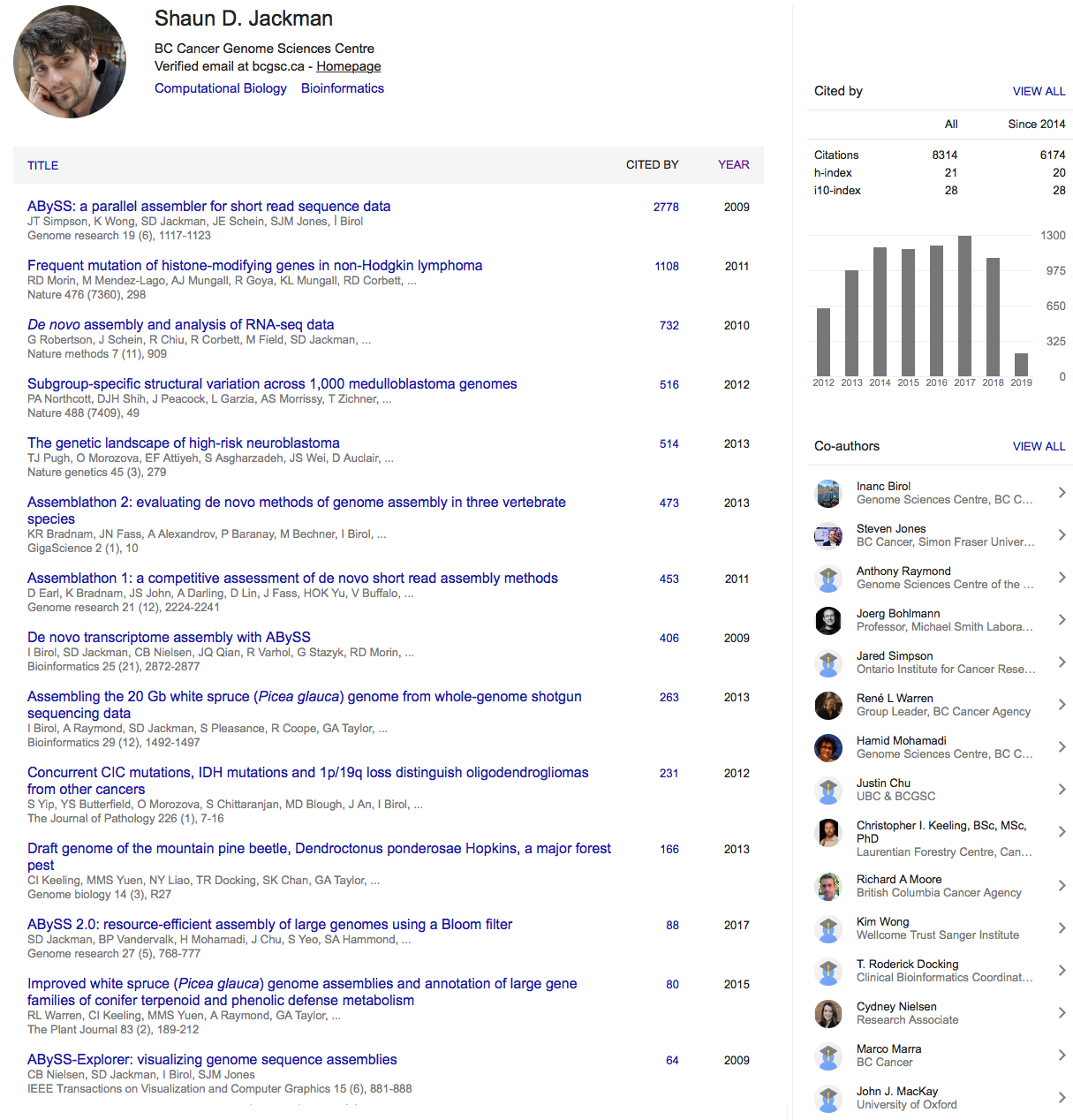
- 12.5 Gbp genome size estimated by flow cytometry
(Hizume et al. 2001 doi.org/d89svf) - 9.8 Gbp genome size estimated by GenomeScope
- 8.0 Gbp assembled in scaffolds 1 kbp or larger
Western Redcedar BUSCO
60.4% of core single-copy genes present (BUSCO)
- 53.9% complete
- 6.5% fragmented
- 39.6% missing
Physlr
Physical Maps of Linked Reads