Results
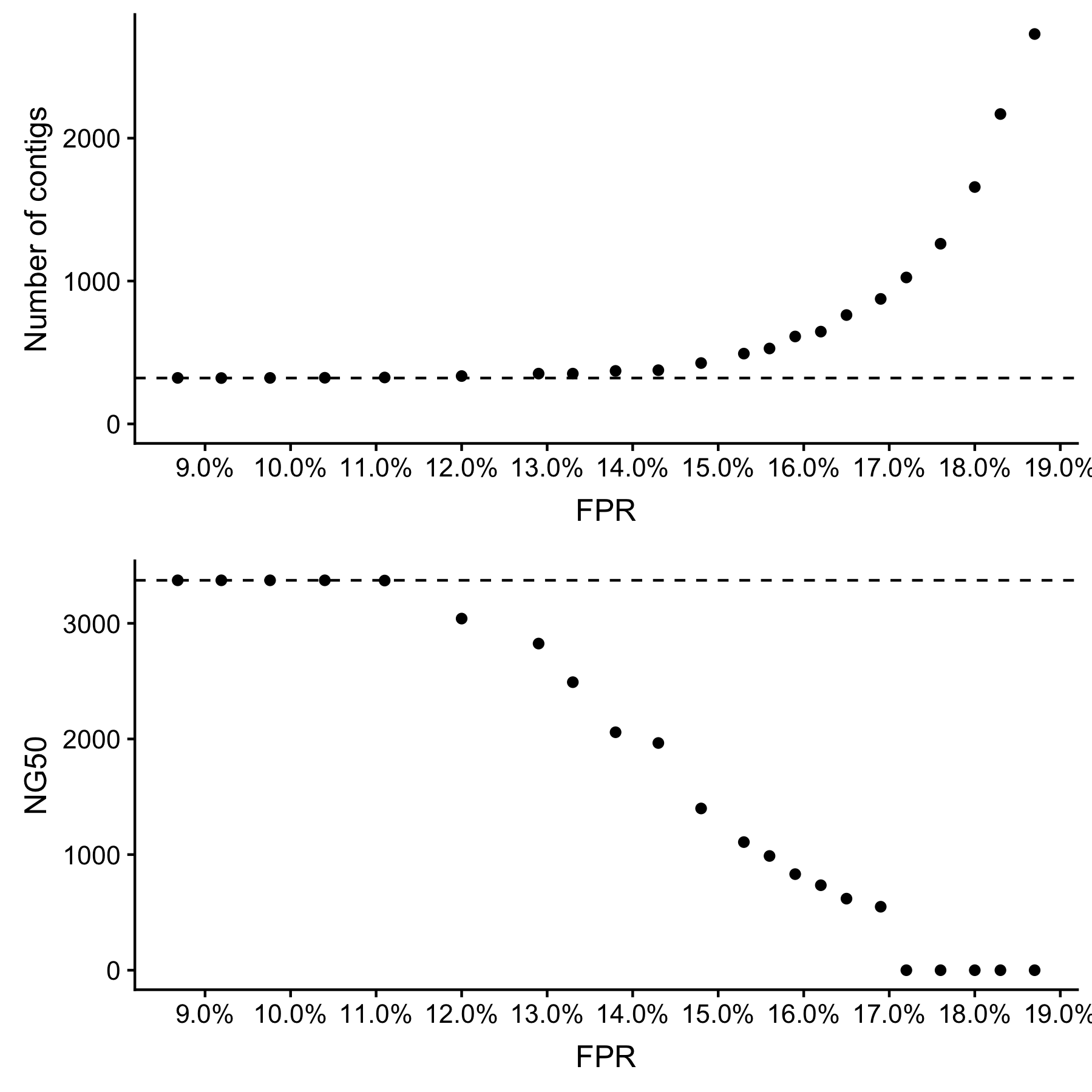
Assembly performance is unaffected so long as the FPR is below 11%. We see degraded assembly performance with FPR between 11% and 17%, and severely degraded assembly performance above 17% (Fig. 1). This result is consistent with Pell et al. (2012), who predict that the graph structure will degrade quickly with a FPR above 18.3%.
Fig. 1: Assembly performance is degraded when the FPR is above 11%, and severely degraded above 17%.
| FPR | Contigs | NG50 (bp) |
|---|---|---|
| 8.7% | 322 | 3,372 |
| 9.2% | 321 | 3,372 |
| 9.8% | 322 | 3,372 |
| 10.4% | 323 | 3,372 |
| 11.1% | 325 | 3,370 |
| 12.0% | 335 | 3,041 |
| 12.9% | 352 | 2,825 |
| 13.3% | 352 | 2,491 |
| 13.8% | 371 | 2,058 |
| 14.3% | 376 | 1,965 |
| 14.8% | 426 | 1,399 |
| 15.3% | 492 | 1,108 |
| 15.6% | 528 | 988 |
| 15.9% | 612 | 831 |
| 16.2% | 646 | 735 |
| 16.5% | 762 | 619 |
| 16.9% | 875 | 549 |
| 17.2% | 1,025 | 0 |
| 17.6% | 1,261 | 0 |
| 18.0% | 1,658 | 0 |
| 18.3% | 2,169 | 0 |
| 18.7% | 2,729 | 0 |
Methods
- ABySS 2.1.4
- Makefile
- Sequencing data
References
Pell, J., Hintze, A., Canino-Koning, R., Howe, A., Tiedje, J. M., & Brown, C. T. (2012). Scaling metagenome sequence assembly with probabilistic de Bruijn graphs. Proceedings of the National Academy of Sciences, 109(33), 13272–13277. https://doi.org/10.1073/pnas.1121464109