The ORCA bioinformatics environment is a Docker image that contains hundreds of bioinformatics tools and their dependencies. The ORCA image and accompanying server infrastructure provide a comprehensive bioinformatics environment for education and research. The ORCA environment on a server is implemented using Docker containers, but without requiring users to interact directly with Docker, suitable for novices who may not yet have familiarity with managing containers. ORCA has been used successfully to provide a private bioinformatics environment to external collaborators at a large genome institute, for teaching an undergraduate class on bioinformatics targeted at biologists, and to provide a ready-to-go bioinformatics suite for a hackathon. Using ORCA eliminates time that would be spent debugging software installation issues, so that time may be better spent on education and research.
Paper doi.org/c4mw · Docker Image bcgsc/orca · Source code on GitHub
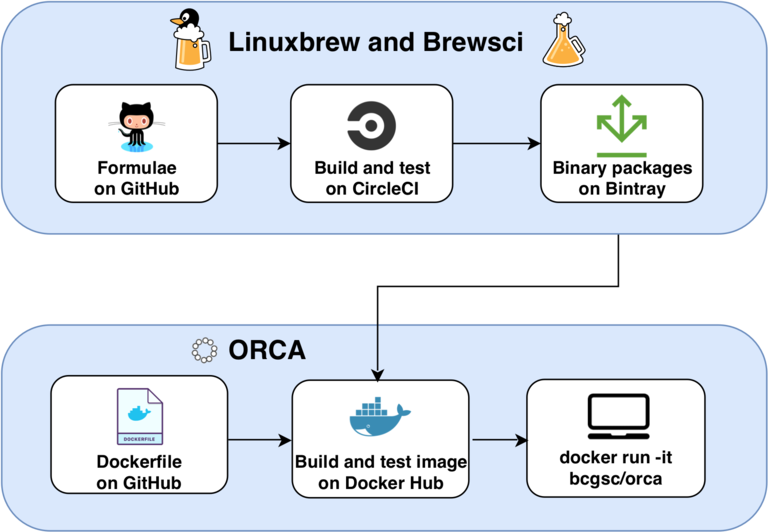
The architecture of ORCA. The package scripts of Homebrew and Brewsci, called formulae, are stored on Github. The precompiled binary packages of Homebrew are built and tested on CircleCI and stored on Bintray. The ORCA Dockerfile is stored on GitHub. The ORCA Docker image is built, tested, and stored on Docker Hub. The system administrator or user pulls the image from Docker Hub to run on their server or workstation.